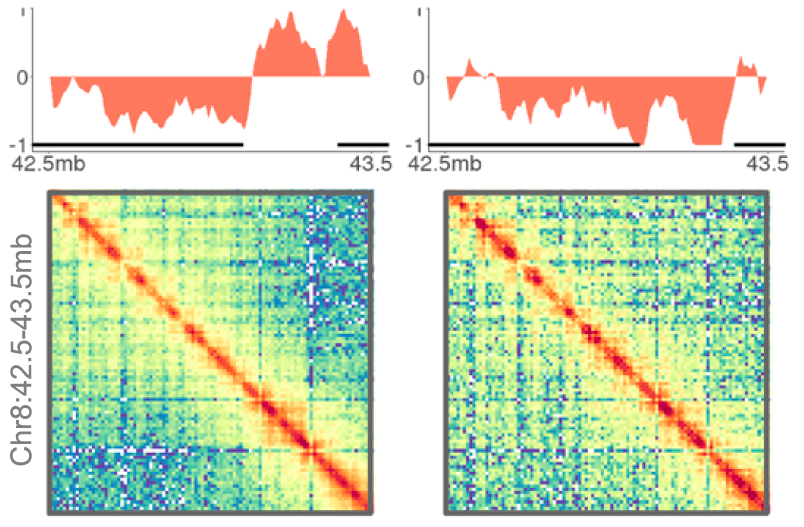
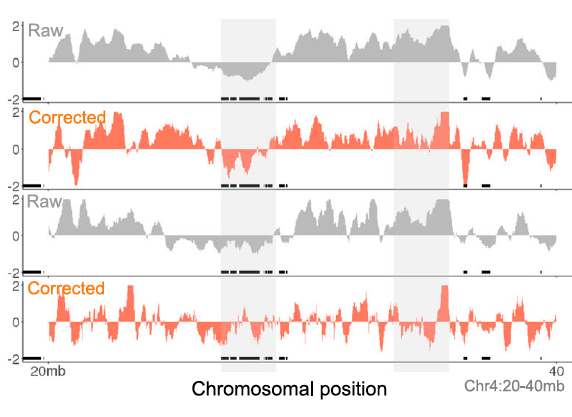
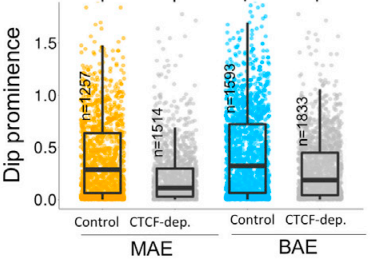
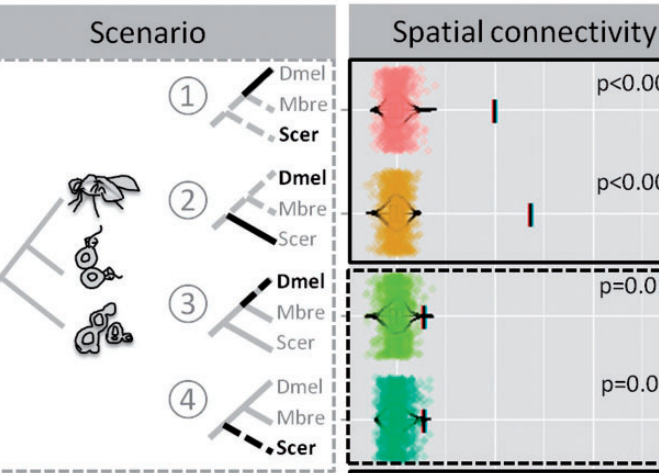
Computational biology is an inevitable and intergral part of modern day biology. We have rigorous expertise in computational genomics and computational structural biology at our institute. Dr. Kuljeet Sandhu's lab primarily addresses the questions related to structural, functional, and evolutionary dynamics of linear and three-dimensional genome organisation of eukaryotic genomes. The genomic basis of many lineage specific phenotypes in vertebrates remains poorly understood. His group has demonstrated that the structural reorganization of genome may explain evolution of certain phenotypes. In particular, they have shown how genome organization profoundly implicates in loss of brain related traits and gain of cancer resistance in certain rodent clades. They have also demonstrated how the CTCF mediated three dimensional genome organization may have impliacted in the evolution of allele-specific expression of genes in mammals.
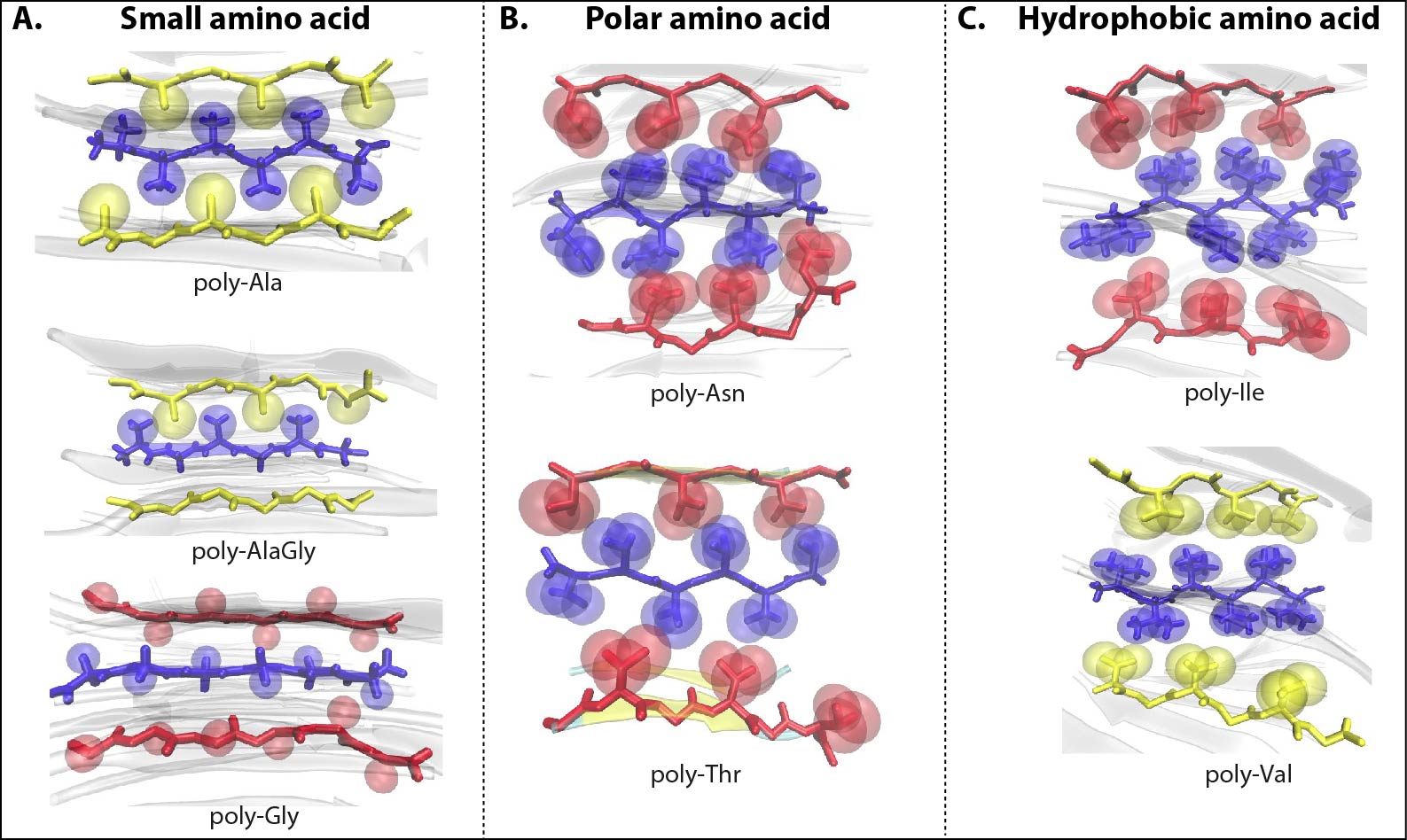
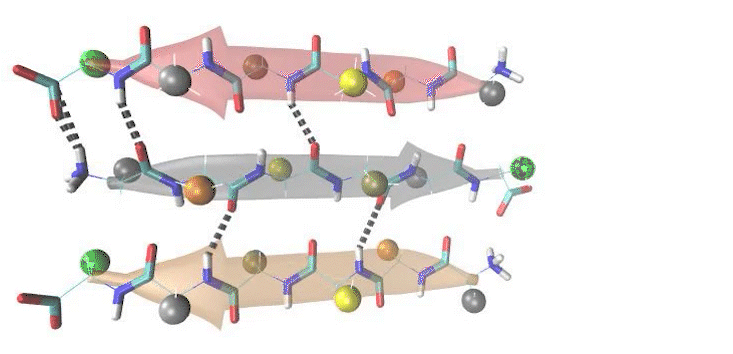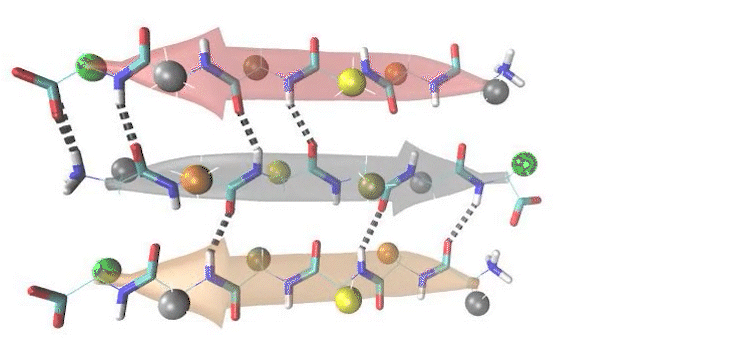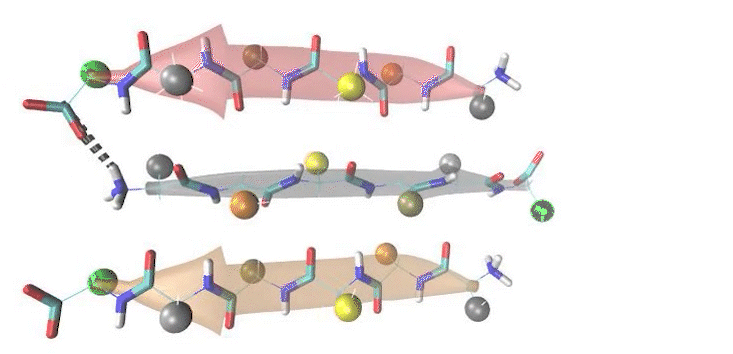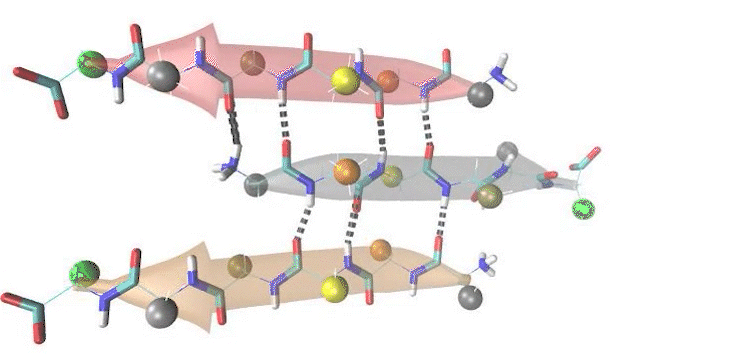
Dr. Shashi Pandit's research vision is to understand the role of protein structures in shaping the molecular evolution of protein function(s). His group is engaged in addressing various biologically interesting questions such as to understand the mechanistic basis of enzyme promiscuity, relating spider silk sequences to their mechanical features, single residue mutational affect(s) on allosteric behaviour of proteins, proteome diversity due to alternative splicing, and how intra-domain motions modulate function of proteins. To study the tensile strength of silk fibre, they have used nanocrystal rupture study by constant velocity pulling SMD simulations and determined ultimate tensile strength of modeled silk nanocrystals. They modeled homopolymer consisting of amino sequences. Based on physiochemical properties the models are cataegorized into Small Amino acid models: poly(Ala), poly(Ala-Gly), poly(Gly); Hydrophobic amino acid models: poly(Ile), poly(Val) and Polar amino acid models: poly(Thr) and poly(Asn). They investigated the modeled nanocrystals for amino acid sequences for packing of side chain and geometry of nanocrystals. In general, (see figure below) the packing remains similar among models. However, nanocrystals adopt twisted cubodial geometry for bulky amino acid side chains. Further, the constant velocity pulling SMD simulations revealed that the natually occuring sequence motids of (Ala/Ala-Gly) have superior mechanical properties than other modeled sequence motifs. Surprisingly, the enhanced side-chain interactions in homo(poly)-polar/hydrophobic amino acid models are unable to augment backbone hydrogen bond cooperativity to increase mechanical strength.